Research
Homeostatic Regulation in Neural Circuits
Most neural networks perform their function reliably throughout an animal’s life, in spite of on-going molecular turn-over and developmental and environmental changes. This is most evident in central pattern generators, i.e. circuits that generate the rhythmic motor patterns that underlie periodic (and often vital) behaviors such as breathing, chewing, walking, and swimming.
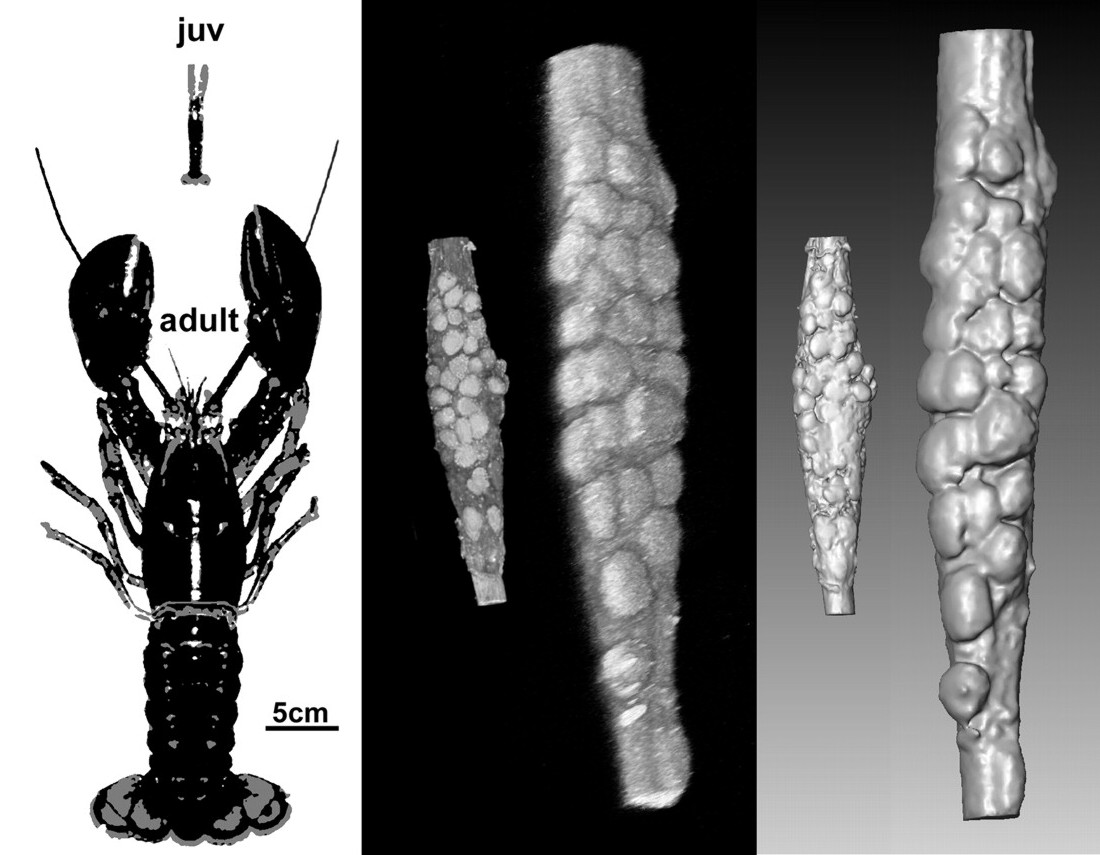
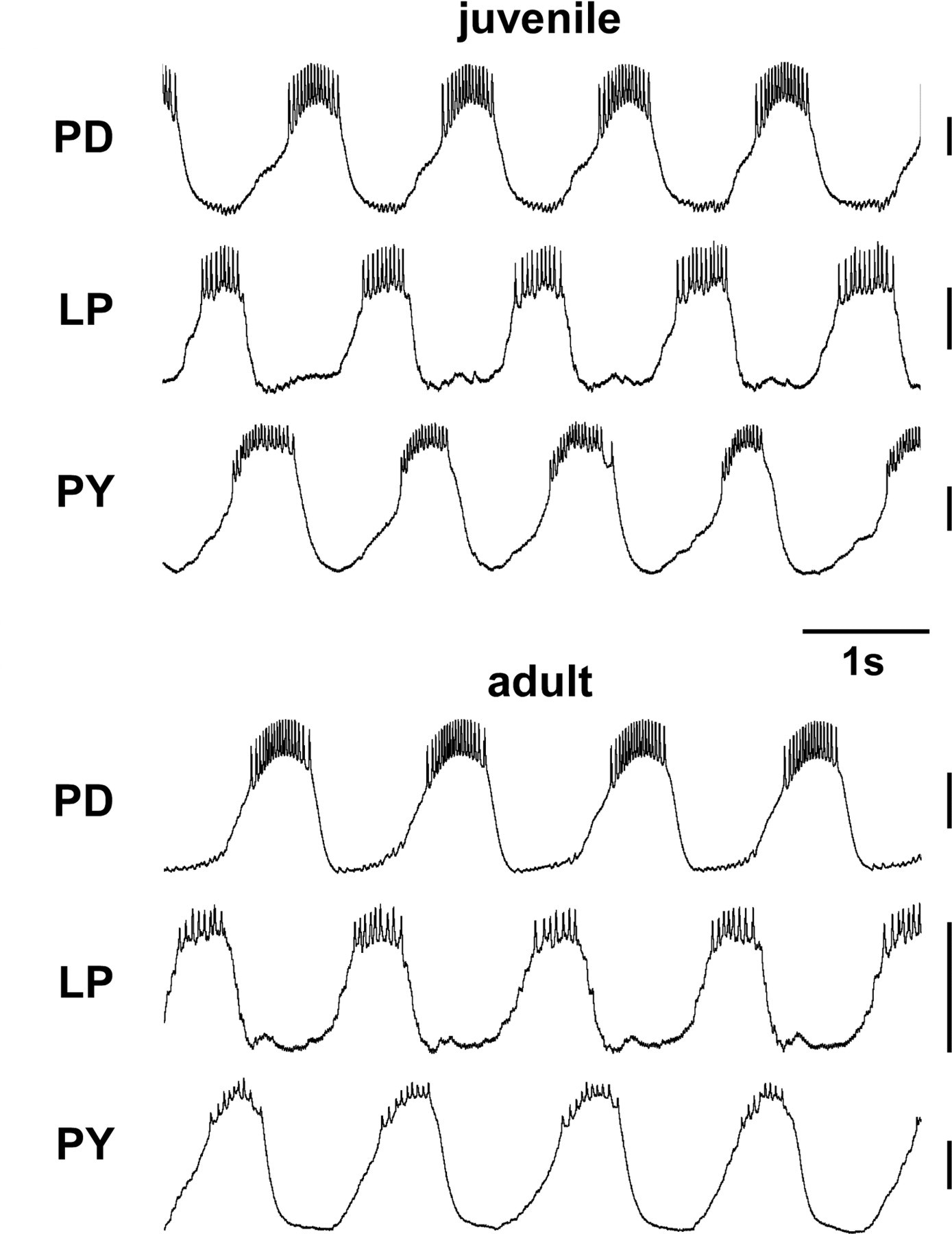
The pyloric pattern generating circuit in the lobster stomatogastric ganglion reliably generates very similar motor rhythms throughout development (right), even though the animal (left) and the stomatogastric ganglion and its neurons (middle) grow dramatically. The volume renderings and surface reconstructions from confocal scans of ganglion autofluorescence show a juvenile and an adult ganglion at the same scale, with individual stomatogastric neurons visible. From Bucher, Prinz, Marder (2005).
Our aim is to understand what classes of regulatory mechanisms underlie functional stability in neuronal circuits, and we approach this question with a computational method that examines many different regulatory pathway architectures in a model network and identifies those that support homeostatic regulation in the face of a variety of perturbations.
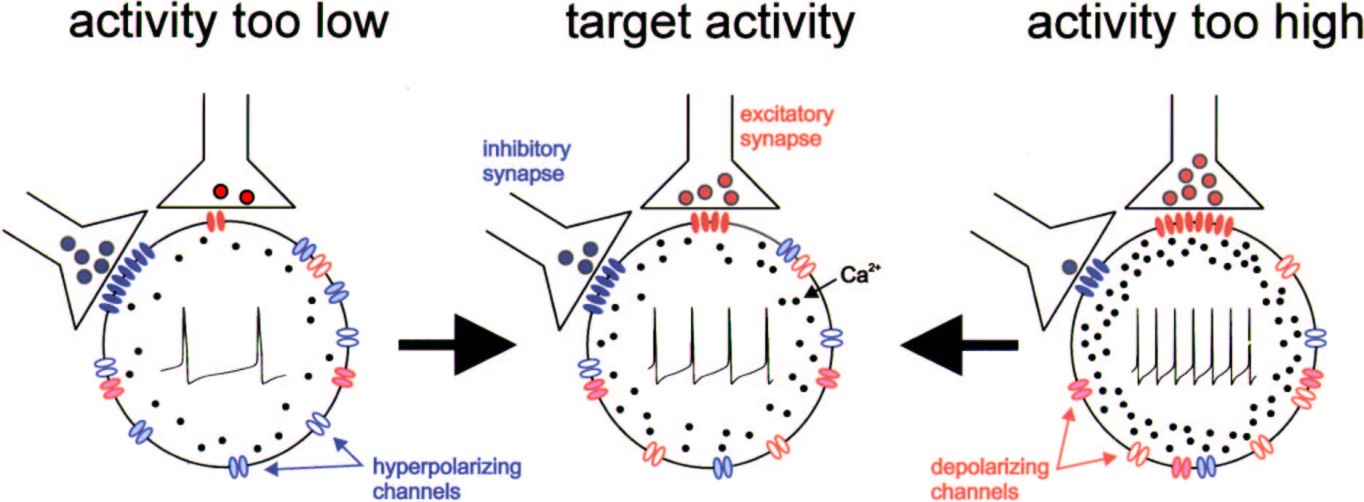
Schematic showing the basic concept of one possible homeostatic regulation mechanism, activity-dependent regulation of intrinsic and synaptic properties. In this paradigm, a neuron senses its own activity by monitoring intracellular calcium concentration, which is closely tied to the electrical activity level of the cell. If activity falls below a target level, the cell reacts to the resulting decrease in intracellular calcium concentration by up-regulating excitatory membrane currents and synapses and down-regulating inhibitory currents and synapses, which brings the activity back to the target level. The same mechanisms act in the opposite direction if activity is too high. From Marder, Prinz (2002).
High-Dimensional Model Databases
A crucial step towards understanding homeostatic regulation is the identification of parameter constellations that allow a neuronal network to generate functional output – a parameter in this context can be any property that influences network behavior, such as the density of a given type of ion channel in a neuron’s membrane or the strength of a particular synapse in the circuit. Previous work resulted in a computational method that assembles information about the behavior of individual model neurons or networks with millions of different parameter combinations in databases (such as the one described here) that can then be analyzed to identify parameter constellations that generate functional behavior. This work has demonstrated that similar, functional behavior can result from very different and widely varying parameter constellations both at the cellular and at the network level. This suggests that homologous neural circuits in different animals, or the same circuit in the same animal at different times, can arrive at very different solutions for generating the same functional behavior, implying that the task of homeostatic regulation may be less daunting than initially thought. We are currently developing this line of work in two different directions: One direction is the application of the approach of database construction and analysis to a number of other neurobiological questions related to homeostasis. In this direction, we studied possible ways calcium can be used as an activity sensor for homeostasis. Using a simulation study, we found optimal parameters and localizations for calcium sensing in the pyloric network (Gunay and Prinz, 2010). The other direction, in collaboration with the labs of Tim Hickey and Eve Marder at Brandeis University, is the development of visualization and analysis tools that will facilitate mining the wealth of information contained in the high-dimensional datasets resulting from this approach.
Synchronization of Neural Oscillators
A separate line of work, in collaboration with Carmen Canavier at the University of New Orleans and Rob Butera at Georgia Tech, uses hybrid network methods that couple model neurons to biological neurons with the help of the dynamic clamp. This project studies the way in which oscillatory – i.e. spiking or bursting – neurons interact in a network setting to produce the periodic motor patterns that underlie many crucial repetitive behaviors.